Document Type : Original Research Article
Authors
1 Plant Biotechnology Department, College of Biotechnology, Al- Nahrain University, Baghdad, Iraq
2 Chemistry Department, College of Science, Al- Nahrain University, Baghdad, Iraq
Abstract
This study focuses on synthesizing three pyrazole derivatives, namely B, C, and D. Fourier transform infrared spectroscopy (FTIR) and proton nuclear magnetic resonance spectroscopy (1H-NMR) were used to study the synthesized derivatives. Pyrazole derivative B was produced from the chalcone derivative (A) reaction with (4-nitrophenyl) hydrazine. The pyrazole derivatives C and D were produced from the reaction of chalcone derivative (A) with hydrazide derivatives (N-(2-oxopropyl)benzamide and 3-acetyl-1-cyclopropyl-6-fluoro-7-(piperazine-1-yl)quinolin-4(1H)-one), respectively. The Purification of α-amylase was conducted on pancreatic cancer patients from Iraq, employing three distinct purifying techniques. The experiment yielded a high level of enzyme activity, specifically 7 U/mL and a specific activity of 8.75 U/mg protein. These results were achieved using an ammonium sulfate saturation ratio of 65%. The graph shows a peak in enzyme activity at 3.75 U/mL in the elution area. This peak, fraction 55, has 11.3 U/mg protein activity. The stability of α-amylase was constant over a pH range of 5.0 to 9.0. In the pH range of 6 to 7, enzyme stability is highest at pH 7. At pH 5 and 9, strength decreased. The enzyme was less active at pH 5 and 6 but more active at pH 7 and 9. However, enzyme activity peaked at pH 8.0. The α-amylase enzyme maintained 100% activity at 27-37 °C, demonstrating stability. However, enzyme activity decreased to 50% at 47 °C when temperature increased. The inhibitory effect of bis-chalcone and pyrazole derivatives increased with concentration. The results show that compound B has the most significant inhibitory efficacy, at 50%.
Graphical Abstract
Keywords
Main Subjects
Introduction
The pancreatic enzyme known as alpha-amylase, also referred to as a-1,4-glucan glucanohydrolase (EC 3.2.1.1) [1], has been the subject of significant research in several species, such as rats, mice, rabbits, humans, dogs, and guinea pigs [2]. Many investigators have confirmed the presence of many electrophoretic and immunological variations of a-amylase [3]. For instance, alpha-amylase is a prominent component of pancreatic and parotid secretions [4]. In addition, the salivary glands produce and release it in small amounts [4, 5]. However, these enzymes display variations in their peptide makeup [6]. Ac-amylase is a good indicator for studying how differential gene expression is controlled in pancreatic acinar cell neoplasia [7] because it is easy to find and purify and contains isoenzymes.
The pyrazole derivatives belong to a unique group of N-heterocyclic compounds (NHCps) defined by a heteroaromatic five-membered ring structure with two nitrogen atoms next to each other [8]. This annular structure consists of a pyrrole-type nitrogen atom, which acts as a proton donor, and a pyridine-type nitrogen atom, which acts as a proton acceptor [9]. Pyrazoles can have mild acidic or essential properties, and the strength of these properties depends a lot on the substituent groups attached to their molecular structure. The remaining three positions within the ring allow for structural modifications either through the utilization of suitable precursors or through post-functionalization reactions after the formation of the pyrazole ring [10]. These modifications result in many synthetic, biological, and photophysical properties for pyrazoles [11]. Numerous pertinent examples [12] show that these variations allow for the formation of more complex structures.
In this study, novel pyrazole (B-D) derivatives have been synthesized using previous chalcones as foundation structures. The application of various spectroscopic techniques has determined the forms of all the synthesized products. Furthermore, the synthesized compounds were screened against α amylase and assessed for their potential anti-pancreatic cancer activities using the MTT procedure.
Experimental
Materials and methods
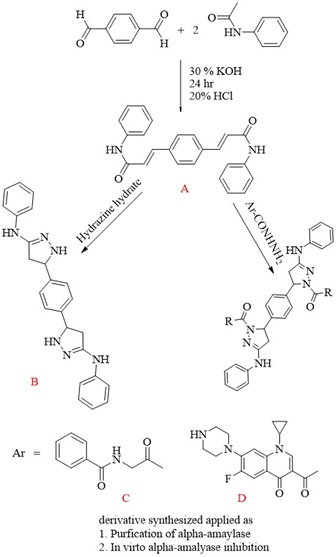
Synthesis of 3,3'-(1,4-phenylene) bis (N-phenyl acrylamide) (Bis-chalcone A)
A 0.036 mole of acetanilide and a 0.018 mole of terephthaldehyde in an ethanolic solution of 30% KOH were stirred overnight at room temperature. The solution was kept in the refrigerator for two hours, poured into ice-cold water, and acidified with 20% diluted hydrochloric acid. The precipitate separated recrystallized from ethanol [13,14]. The precipitate is pale yellow; M.wt: 368.43; m.p: 214 °C; yield: 86%.
Synthesis of 5,5'-(1,4-phenylene)bis(1-(4-nitrophenyl)-N-phenyl- 4,5-dihydro-1H-pyrazol-3-amine) (Pyrazole derivative B)
A 0.003 mole of chalcone derivative (A) was refluxed with 0.02 mole of (4-nitrophenyl) hydrazine in 20 mL of absolute ethanol at 90 °C for 11 hours. After cooling the mixture, it was washed with water several times, filtered, dried, and recrystallized in ethanol [15]. The derivative (B) is brown, has a molecular weight of 638.67, a melting point of 310 ˚C, and a yield of 71%.
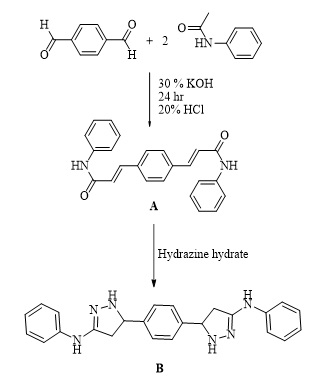
Scheme 1. Synthesis of derivatives (B).
Synthesis of N,N'-((5,5'-(1,4-phenylene)bis(3-(phenylamino)-4,5-dihydro-1H-pyrazole-5,1-diyl))bis(2-oxoethane-2,1-diyl))dibenzamide (Pyrazole derivative C) and synthesis of 3,3'-(5,5'-(1,4-phenylene)bis(3-(phenylamino)-4,5-dihydro-1H- pyrazole-1,1'-carbonyl))bis(1-cyclopropyl-6-fluoro-7-(piperazine-1-yl)-quinoline- 4(1H)-one) (Pyrazole derivative D)
A 0.003 mole of ciprofloxacin, a 0.005 mole of hippuric acid, 30 mL of absolute methanol, and 1 mL of sulfuric acid were refluxed for 6 hours. After cooling the mixture, it was washed with water several times, filtered, dried, and recrystallized with ethanol. 0.005 moles of hydrazide derivatives (N-(2-oxopropyl)benzamide and 3-acetyl-1-cyclopropyl-6-fluoro-7-(piperazine-1-yl) quinoline-4 (1H)-one), respectively, were added to 3 mL of hydrazine hydrate (80%) and refluxed for four hours, and then 5 mL of ethanol was added and re-condensed for 1 hour. After cooling, the product was filtered. Recrystallized from ethanol [16].
The pyrazole derivative (C) is white in color, weighs 718.80 grams, has a melting point of 334 °C, and yields 77%. The pyrazole derivative (D) color is off white; M.wt. is 1023.14; m.p. is 292 °C; yield is 69%.
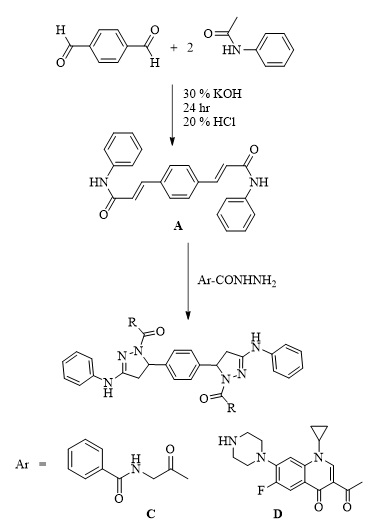
Scheme 2. Synthesis of derivatives (C and D).
Results and Discussion
3,3'-(1,4-phenylene) bis (N-phenylacrylamide) (Bis-chalcone) A, molecular formula: C24H20N2O2; colour: Pale Yellow; M.wt: 368.43; m.p: 214 °C; yield: 86%. The FT-IR spectra of pyrazole (B) peaked at (3427) cm-1, which belongs to the N-H group. Another peak belongs to the C=C aliphatic that appeared at 1598 cm-1. The carbonyl group showed at 1664 cm-1.
5,5'-(1,4-phenylene)bis(1-(4-nitrophenyl)-N-phenyl- 4,5-dihydro-1H-pyrazol-3-amine) (B), molecular formula: C36H30N8O4; color: Brown; M.wt: 638.67; m.p: 310 °C; yield: 71%. The FT-IR spectra of pyrazole (B) peaked at (3398) cm-1, which belongs to the N-H group. Another peak belongs to C=C Aliphatic, which appeared at 1629 cm-1. The imine group showed at 1650 cm-1. 1HNMR (Solvent: DMSO; PPM), Multiplet 24H of aromatic rings (6.75- 8.27), Singlet 2H of amine groups (10.01), Doublet 4H of CH2 in pyrazole rings (2.00), Triplet 2H of –CH in pyrazole rings (3.73), and 2H free of NH (4.50) [17]. N,N'-((5,5'-(1,4-phenylene)bis(3-(phenylamino)-4,5-dihydro-1H-pyrazole-5,1-diyl))bis(2-oxoethane-2,1-diyl))dibenzamide (C), molecular formula: C42H38N8O4; color: White; M.wt: 718.80; m.p: 334 °C; yield: 77%. The FT-IR spectra of pyrazole (B) showed a peak at (3369) cm-1, which belongs to the N-H group [18]. Another peak belongs to C=C, which appeared at 1542 cm-1. The imine group showed at 1635 cm-1. 1HNMR (Solvent: DMSO; PPM), Multiplet 24H of aromatic rings (7.01- 8.62), Singlet 4H of amine groups (9.90 – 9.99), Doublet 4H of CH2 in pyrazole rings (1.81), Triplet 2H of –CH in pyrazole rings (3.74), and Singlet 2H free of NH (4.49). 3,3'-(5,5'-(1,4-phenylene)bis(3-(phenylamino)-4,5-dihydro-1H- pyrazole-1,1'-carbonyl))bis(1-cyclopropyl-6-fluoro-7-(piperazin-1-yl)-quinolin- 4(1H)-one) (D), molecular formula: C58H56F2N12O4; color: Off White; M.wt: 1023.14; m.p: 292 °C; yield: 69%. The FT-IR spectra of pyrazole (B) peaked at (3296) cm-1, which belongs to the N-H group. Another peak belongs to C=C, which appeared at 1600 cm-1 [19]. The imine group showed at 1632 cm-1. 1H-NMR (Solvent: DMSO; PPM), Multiplet 24H of aromatic rings (7.01- 8.06), Singlet 4H of amine groups (9.96) [20], Doublet 4H of CH2 in pyrazole rings (2.05-2.10), Triplet 2H of –CH in pyrazole rings (4.59), Singlet 2H free of NH (4.49), and Quartet 8H of CH2 in cyclopropane [21]. Purification of α-Amylase: AMY was purified from Iraqi pancreatic cancer patients using three purification methods: precipitation by ammonium sulfate ion-exchange chromatography and gel filtration chromatography [22]. The description of each step is as follows:
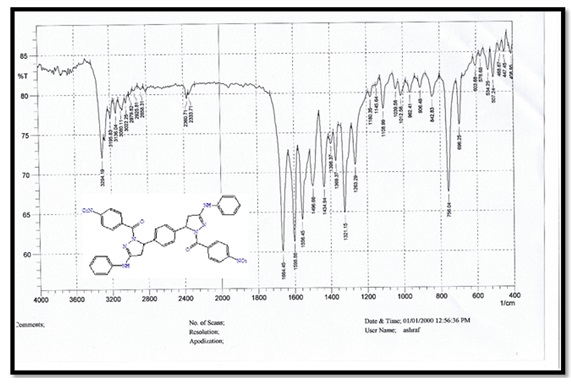
Figure 1. FTIR spectrum for compound A.
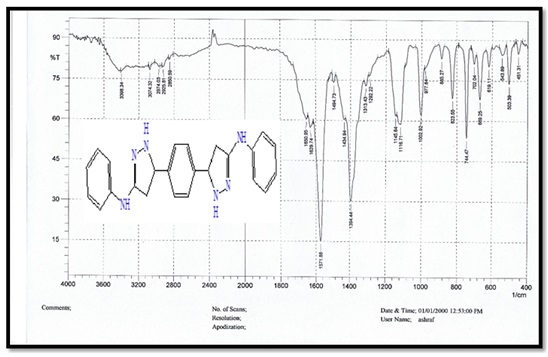
Figure 2. FTIR spectrum for compound B.
Ammonium Sulfate Precipitation: Salting out, which depends on ammonium sulfate salt precipitation due to its high solubility and affordable method for protein purification, is one of the most often used processes in enzyme purification.
In theory, the ammonium and sulfate ions draw the water molecules between protein chains, which causes the proteins (including the enzyme of interest, AMY) to precipitate out of the solution. Results illustrated in Table 1 show that the high enzyme activity (7 U/mL and specific activity (8.75 U/mg protein)) were obtained in an ammonium sulfate saturation ratio of 65%.
After ammonium sulfate precipitation, dialysis was performed in order to neutralize the enzyme crude using 7000 cutoff dialysis against potassium phosphate buffer [23].
Characterization of Purified α-Amylase
1- Effect of pH Stability on α-Amylase Enzyme
Enzymes are substances that are affected by changing the pH value depending on the nature of their catalytic group and the enzyme source. α - Amylase was stable across an extensive pH range (5.0-9.0). The optimal pH for enzyme stability is between 6 and 7, with the highest strength at 7 and lower stability at pH 5 and 9, respectively (Figure 8).
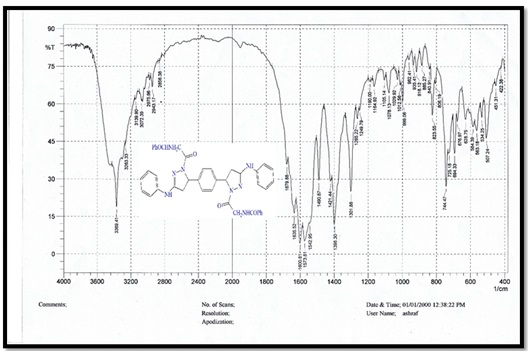
Figure 3. FTIR spectrum for compound C
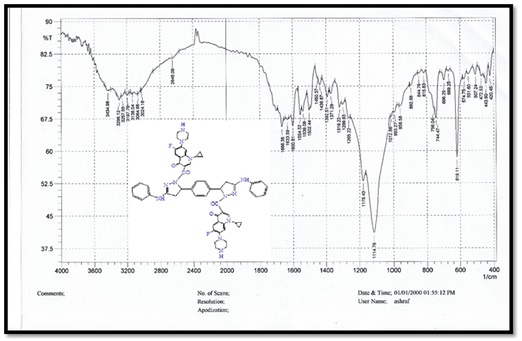
Figure 4. FTIR spectrum for compound D
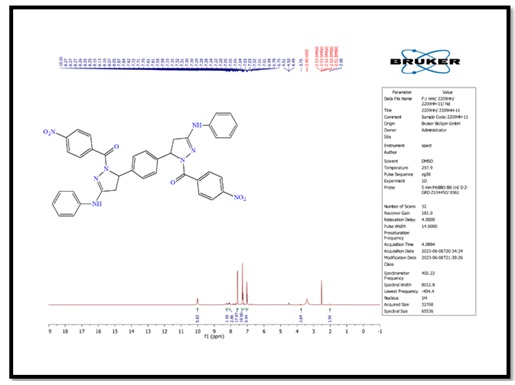
Figure 5. 1H-NMR spectrum of compound (B)
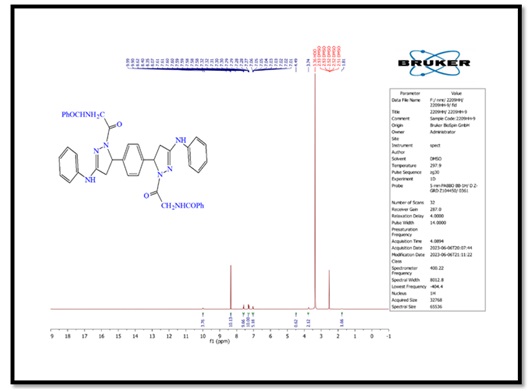
Figure 6. 1H-NMR spectrum of compound (C)
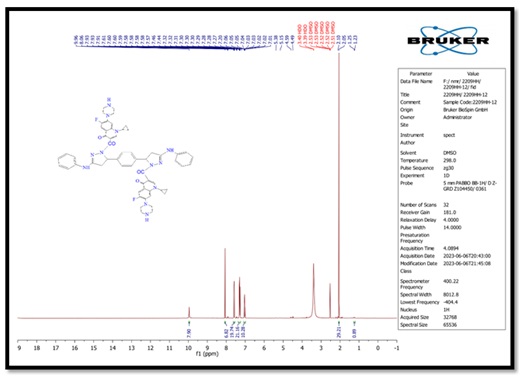
Figure 7. 1H-NMR spectrum of compound (D)
Table 1. Purification steps of serum α-Amylase
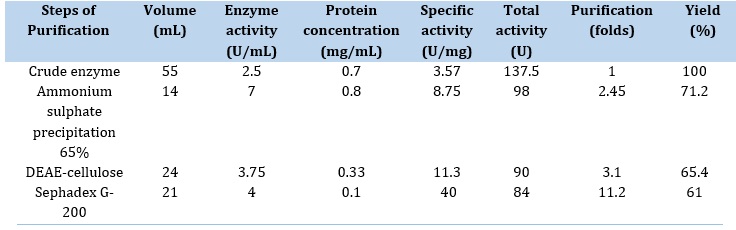
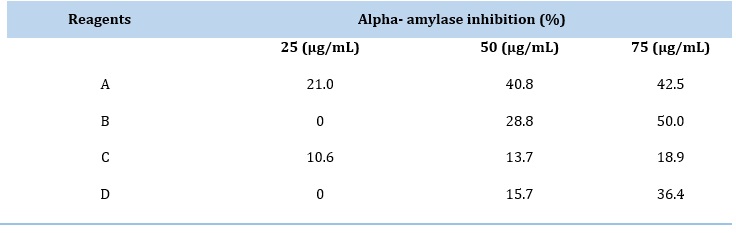
Table 2. The inhibition efficiency of purified α-Amylase
According to these findings, α-Amylase tolerates both acidic and alkaline environments, but prefers the neutral. An enzyme's conformation can change due to the pH, and the ionization groups in the enzyme can also be affected, either the groups in the substrate or the enzyme's pocket [24].
Effect of Temperature Stability on α-Amylase Enzyme
An essential element influencing enzyme activity is temperature. Alpha-amylase was incubated at a range of temperatures between (27 and 47 °C), and the amount of still detectable activity was calculated using an enzyme activity assay to investigate how temperature affects the stability of the enzyme. The result shows that α-Amylase was stable and maintained its activity 100% at (27-37) °C; when the temperature was increased, the remaining activity began to decrease to 40% at 47 °C. Our results agreed with Rehab et al.'s study, which showed that α-Amylase was stable and preserved 100% activity at 37 °C [25].
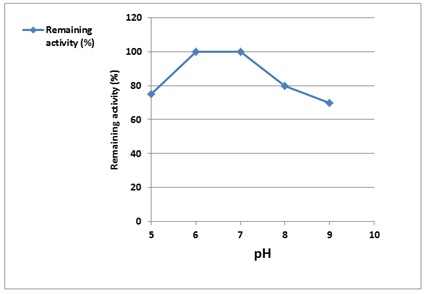
Figure 8. Effect of pH on purified α-Amylase activity enzymes are substances that are affected by changing the pH value depending on the nature of its catalytic group and the enzyme source
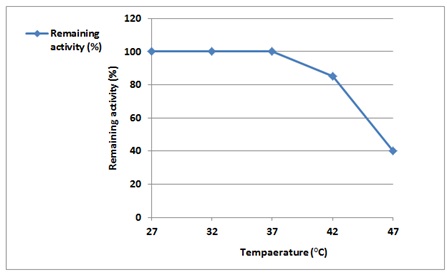
Figure 9. Effect of temperature on purified α-Amylase activity
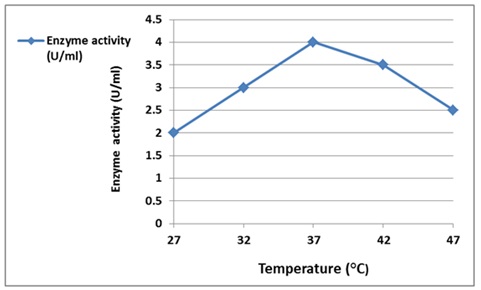
Figure 10. Effect of temperature activity on α-Amylase enzyme
Effect of Temperature Activity on α-Amylase Enzyme
Temperature is an essential factor which affects enzyme activity. The effect of temperature on purified alpha-amylase activity is presented in Figure 10. The highest enzyme activity was recorded at 37 ˚C with an observed decrease in action if the incubation temperature increased or decreased. The results showed an increase in enzyme reaction activity until it reached 37 ˚C, and then it began to decline.
The increase in temperature resulted in imparting more kinetic energy to the reactant molecules, according to results showed by Spier, M.R. et al., in more productive collisions per unit of time, but that should be within the intact and proper configuration of the tertiary structure of an enzyme maintained primarily by a large number of weak non-covalent bonds, so if the molecule absorbs too much energy, the tertiary structure will disrupt, and the enzyme will be denatured, that is, loss of catalytic activity.
In Vitro α-Amylase Inhibition
Inhibitors α-amylase are also known as starch blockers because they do this by inhibiting the hydrolysis of 1,4-glycosidic bonds in starch and other oligosaccharides into maltose and other simple sugars, which slows or stops the body from absorbing starch. The bis-chalcone and pyrazole derivatives were in vitro assayed for their potent inhibition activity against purified α-Amylase from sera of pancreatic cancer patients.
The inhibitors were prepared in different concentrations (25, 50, and 75) μg/mL, and their inhibition data are listed in Table 2 and Figure 10. In general, the inhibitory activity of bis-chalcone and derivatives was increased with increased concentration. Based on the results, compound (B) is the most potent inhibitor, with a maximum inhibition of 50 % [26].
Conclusion
The characterization of all produced compounds (A–D) was conducted using Fourier transform infrared spectroscopy (FTIR) and proton nuclear magnetic resonance spectroscopy (1H-NMR). Three approaches were employed to separate α-amylase from pancreatic cancer patients in Iraq. The study resulted in a high level of enzyme activity, measuring 7 U/mL and a specific activity of 8.75 U/mg of protein. These values were obtained when the ammonium sulfate saturation level reached 65%. Within the elution zone, the enzyme exhibits its highest activity level at a concentration of 3.75 U/mL, accompanied by a specific activity of 11.3 U/mg protein, which occurs at fraction number 55. The activity of α-amylase remained consistent within the pH range of 5.0 to 9.0. The optimal pH range for maintaining enzyme stability is typically observed between 6 and 7, with the highest level of stability occurring at a pH of 7.
Conversely, enzyme stability decreases at pH levels between 5 and 9. The activity exhibited a decline at pH levels 5 and 6, while it demonstrated a rise from pH 7 to 9, reaching its peak at pH 8.0. The activity of α-amylase remained constant at 100% within the temperature range of 27–37 °C. However, as the temperature increased to 47 °C, the activity decreased to 50 %. The potency of bis-chalcone (A) and pyrazole derivatives (B, C, and D) in inhibiting a specific action was found to be positively correlated with their concentration. According to the available data, compound (B) has the highest inhibitory potency, with an inhibition rate of 42.5%.
Acknowledgements
We are sincerely grateful to Al-Nahrain University for its support of this work.
Disclosure Statement
No potential conflict of interest was reported by the authors.
Orcid
Dina Naseer Ali : 0009-0007-4464-1466
HOW TO CITE THIS ARTICLE
Dina Naseer Ali*, Ahmed Ahmed, Alaa Hussein J. Al-qaisi. Inhibitory Effect of Newly Prepared Pyrazole Derivatives against Alpha-Amylase in Pancreatic Cancer Cell. Adv. J. Chem. A, 2024, 7(3), 248-259.
DOI: 10.48309/ajca.2024.425005.1447